
|
This document assumes that users have a basic knowledge of the following data formats and the corresponding software packages:
Note: Additional software packages are required to install GrADS. Please see: section 4.2. Installation, below for further information.
In this document we describe how to use GrADS [5] to access HDF files. We use NASA HDF files to show the procedures for accessing or visualizing the data with this program. For the general usages of these packages, please refer to their user's guides, or websites.
One
HDF4 file and two
HDF-EOS2
files were used to demonstrate
how to access or visualize HDF data with these packages. These files
are used to store data from the NASA Special Sensor Microwave/Imager
(SSMI) Sensor
[6]and the Advanced
Microwave Scanning Radiometer for the EOS (AMSR-E) satellite system
[7]. Table 1 shows the
instrument names, formats and other information about these
HDF4/HDF-EOS2 files. We also converted HDF-EOS2 files to netCDF-4
classic model-compliant and netCDF-4-compliant HDF5
file and two
HDF-EOS2
files were used to demonstrate
how to access or visualize HDF data with these packages. These files
are used to store data from the NASA Special Sensor Microwave/Imager
(SSMI) Sensor
[6]and the Advanced
Microwave Scanning Radiometer for the EOS (AMSR-E) satellite system
[7]. Table 1 shows the
instrument names, formats and other information about these
HDF4/HDF-EOS2 files. We also converted HDF-EOS2 files to netCDF-4
classic model-compliant and netCDF-4-compliant HDF5 files. In
this document, we mainly refer to the instrument and the description
of physical variables when specifying a file or a field in a file.
However, the file names may be referred to in the code examples
that appear throughout this document.
files. In
this document, we mainly refer to the instrument and the description
of physical variables when specifying a file or a field in a file.
However, the file names may be referred to in the code examples
that appear throughout this document.
| ||||||||||||||||||||
| Table 1 Sample files |
The first file stores the NASA SSMI ocean wind fields data.
In this document, we will use the two variables u10m and
v10m that represent the U component and the
V component of the wind field. Both variables have
three dimensions: time (from January 2005 to December 2005),
longitude, and latitude.
The file name is
atlas.ssmi.ver02.level3.5_5day.s950103.hdf.
Users can download this file from
here.
The second file (AE_RnGd) stores the NASA AMSR-E data that describes the monthly average rainfall accumulation over ocean and land in July 2007.
The format of the original file is HDF-EOS2. The original file name is
AMSR_E_L3_RainGrid_B05_200707.hdf.
Users can download this file from
here.
We used the HDF4-to-HDF5 Conversion tool[8] to convert the AMSR-E HDF-EOS2 file
to a netCDF-4-compliant HDF5 file. The converted file was renamed to either
AMSR_E_L3_RainGrid_B05_200707.h5 or AMSR_E_L3_RainGrid_B05_200707.nc,
as required by the software package processing it.
Users can download this netCDF-4-compliant HDF5 file from
here.
Since the netCDF-4 classic model[9] has more restrictions
than the general netCDF-4 model, two steps are needed to create a
netCDF-4 classic model–compliant HDF5 file.
The first step is to create a netCDF-4-compliant HDF5 file as addressed in the
previous section. The second step is to follow the procedure explained in
Appendix 5.1.
The file is called AMSR_E_L3_RainGrid_B05_200707_flatten.nc, and users can download
this file from
here.
The third file (AE_SI12) contains several fields that represent brightness temperatures, sea ice concentration, and snow depth over sea ice. This file is also from AMSR-E[10]. Unlike the other two files, the polar stereographic projection is used.
The original file is an HDF-EOS2 file and the filename is
AMSR_E_L3_SeaIce12km_B02_20020619.hdf.
Users can download this file from
here.
To create the netCDF-4 classic model-compliant HDF5 file,
one needs to follow the same procedure described in Section 3.2.3.
The converted file name is AMSR_E_L3_SeaIce12km_B02_20020619_flatten.nc.
Users can download this file from
here.
The Grid Analysis and Display System (
GrADS ) is an interactive desktop
tool that is used for easy access, manipulation, and visualization of
earth science data. In this section we explain how to read and visualize HDF4 and HDF5 files.
) is an interactive desktop
tool that is used for easy access, manipulation, and visualization of
earth science data. In this section we explain how to read and visualize HDF4 and HDF5 files.
You must install GrADS version 2.0.a8 or later
which supports netCDF-4 since the earlier versions of GrADS
do not support netCDF-4
since the earlier versions of GrADS
do not support netCDF-4 Classic files.
Please see:
The complete list of supplementary libraries
required for installing GrADS.
Classic files.
Please see:
The complete list of supplementary libraries
required for installing GrADS.
Alternatively, you can install
OpenGrADS
version 2.0.a5.orga.5 or later which also supports
netCDF-4 Classic files.
Classic files.
Three methods are provided to read
netCDF or HDF4 SDS files:
or HDF4 SDS files:
sdfopen, xdfopen, and open.
On Unix/Linux platforms, please note:
Some environment variables must be set before starting GrADS.
In particular, set $GADDIR to ...src/grads-install/data
is required by the sdfopen method.
After reading a file, data can be manipulated and visualized in
many ways using the display command. One can change visualization settings using
set lon, set lat, set gxout, and so on.
Like NCL , GrADS can both draw plots on the screen and export them to a file.
, GrADS can both draw plots on the screen and export them to a file.
The sdfopen command accepts the actual file name. Then, GrADS
recognizes longitude and latitude from attributes and reads all values from
the data fields. GrADS can recognize missing values from attributes and can
detect date and time from human-readable strings such as
"hours since 1995-01-01 00:00:00".
This method is the most convenient, but the target file should conform to
COARDS conventions[11]. For example, HDF-EOS2 grid files cannot be opened
using sdfopen because the values of longitude and latitude are not explicitly defined.
When a file does not conform to COARDS conventions,
one can use the xdfopen command. This command requires a user-defined
description file. The xdfopen command receives the description file name
as an argument. Note that xdfopen does not receive the actual data file name
because that is specified in the description file.
Figure 1 shows part of a typical description file
used by the xdfopen command to open an HDF-EOS2 grid file.
The first line of the description file specifies the actual HDF-EOS2 file name.
Then, XDEF and YDEF define longitude and latitude, respectively.
Both XDEF and YDEF receive five arguments. The first argument
is the actual dimension name in the file. The HDF-EOS2 grid file does not store
latitude and longitude as variables. It only defines dimension names,
which are normally XDim and YDim followed by a colon and the enclosing grid name.
These names can be easily fetched with hdp, a command-line dumper utility of HDF4,
or HDFView[12]. In this example,
XDim:grid1 is the actual dimension name defined in the file.
The second argument specifies the size of the dimension, and users can easily get this information
from hdp or HDF View . This is explained in
Appendix 5.2.
. This is explained in
Appendix 5.2.
The remaining three arguments, LINEAR 2.5 5, mean that the longitude starts
from 2.5 degrees and the values increase by a step size of 5. Since these arguments are
not explicitly defined in the file, users need to get these values from the projection
code and related attributes inside the HDF-EOS2 file.
If longitude and latitude values are not linear, one needs to use
LEVELS followed by a list of actual values instead of LINEAR.
In addition, using XDEF and YDEF is not proper if longitude and
latitude are two-dimensional. This is explained in Section 4.3.2.4.
The remaining part defines variables. The line starting with var1 defines one variable,
variable1, from a field, var1, in the file. One should be aware that defining
XDEF and YDEF correctly is crucial to drawing correct plots because GrADS
retrieves each element of variable1 from user-defined XDEF and YDEF options.
If a file does not conform to COARDS conventions and xdfopen fails, one can use the
open command. Like xdfopen, open also requires a description file.
The description file is very similar although the description file for open requires more settings.
Since the open command supports the PDEF option, two-dimensional longitude
and latitude can be handled. The detailed explanation and an example are provided in
Section 4.3.2.4.1.
In this section, we explain how to visualize an HDF4 SDS (4.3.2.1), HDF-EOS2 (4.3.2.2, 4.3.2.4) and netCDF-4 classic model-compliant HDF5 (4.3.2.3, 4.3.2.5).
The variable for ocean wind fields from the SSMI instrument has enough metadata for
GrADS to recognize it; it has longitude and latitude dimension scales, unit, and fill value.
To read this file, sdfopen is sufficient.
Since this file has time dimension as well as longitude and latitude, several plots, based on the time dimension, can be animated. The following is a full list of commands that read the file and draw animated plots. When GrADS shows the prompt, type these statements:
set t 1 73
and set looping on are used for animation. The t implies the time dimension,
and the output will display an animation containing 73 plots. The display command
actually draws an animation. Both u10m and v10m are variable names
defined in the file. The last expression, sqrt(…), is optional; GrADS
colorizes the vector according to the value of this field.
Figure 3 shows the first frame of the animation.
As explained in Section 4.3.1.1,
sdfopen cannot be used to visualize HDF-EOS2 grid data.
Users can use the xdfopen command instead.
Figure 4 shows an example of the description file.
As explained in Section 4.3.1.2, HDF-EOS2 files define dimension names.
In this example, XDim:MonthlyRainTotal_GeoGrid is one actual dimension name defined in the file.
OPTIONS YREV indicates that the latitude has the reverse order.
LINEAR -67.5 5 in the YDEF statement means the first data was measured at -67.5
and the next data was measured at -62.5, which means the location goes from south to north.
However, the actual data in the file were measured from north to south, and this option
provides this information.
We named this description file AE_RnGd.xdf. Under the GrADS environment, that file
name is passed as the argument to xdfopen, as in the following example.
set gxout shaded makes GrADS draw a shaded plot. The display command shows a visualization,
which is represented in Figure 6.
Since the
HDF4-to-HDF5 Converter calculates and writes longitude and latitude from HDF-EOS2
projection code and related attributes,
calculates and writes longitude and latitude from HDF-EOS2
projection code and related attributes,
sdfopen can be used, and tedious tasks
specifying XDEF and YDEF can be avoided.
However, the generated file does not conform to the netCDF-4 classic model due to groups.
To conform to the classic model, the generated file should be manually flattened as we show in
Appendix 5.1.
After the modification, sdfopen can directly open the netCDF-4-compliant HDF5 file.
Be aware that the current GrADS release cannot handle a netCDF-4 classic model-compliant
HDF5 file. Check
Appendix
for more details.
If longitude and latitude values cannot be represented as
one-dimensional arrays, XDEF and YDEF are not enough.
Two-dimensional longitude and latitude are supported through the PDEF command.
For the PDEF option, open should be used instead of
sdfopen or xdfopen
as Section 4.3.1.3 explained.
Figure 8 shows an example
of a description file.
The description file has UNDEF, ZDEF, TDEF
that represent undefined values, Z-axis, and time axis, respectively,
because the open command requires these definitions.
In this example, ZDEF and TDEF are meaningless
because the actual data do not have Z-axis and time axis.
The fill value is undefined in the original file. To generate a
plot that can correctly describe the distribution the physical field,
fill value is set to 0.
PDEF BILIN provides a way for GrADS to obtain geolocation
information as lon2d and lat2d do in
NCL example (Section 4.3.3.4).
However, the geolocation information should be stored in an additional
file, and the file name is passed as an argument to PDEF BILIN.
In this example, bilin_file is the name of the additional file.
This additional file specifies the horizontal array indices of the
data field measured at each grid point defined by XDEF and YDEF.
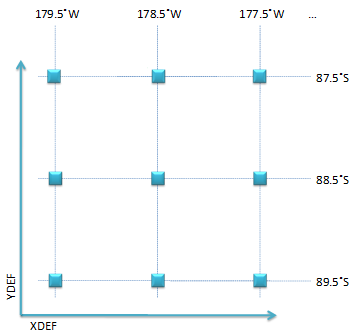
In Figure 8,
XDEF and YDEF define a 360 × 180 horizontal grid.
Shown in Figure 9(a),
each grid point is represented as a rectangle;
the latitude and longitude at each grid point are also specified.
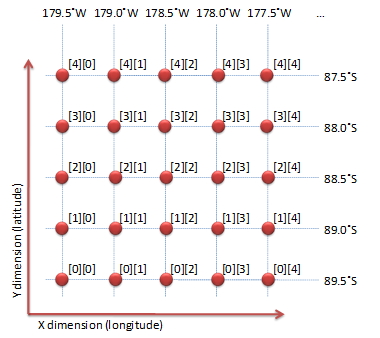
Figure 9(b)
shows the data array index with the corresponding latitude and longitude,
which can be obtained from the EOS2 file. The data array index for each
grid point shown in Figure 9(a)
needs to be mapped from the information shown in
Figure 9(b).
For example, the rectangle at the third column and the second row
(88.5˚S, 177.5˚W) represents the location of the data element of which
the index is [2][4] (Figure 9(b)).
The data array index for each grid point defined by
XDEF and YDEF is then stored in the bilin_file.
GrADS draws the plot based on the array index.
Calculating array indices may not be straightforward for some projections.
If the file is an HDF-EOS2 grid file, however, users can exploit the
HDF-EOS2 API function GDll2ij. This function accepts the
projection code, related parameters, and longitude and latitude,
and it returns indices for the X dimension and Y dimension corresponding
to the given longitude and latitude. Among the API inputs,
the projection code and related parameters are defined in the HDF-EOS2 file.
Longitude and latitude are defined by XDEF and YDEF
in the description file. Given the XDEF and YDEF defined in
Figure 8,
Figure 10,
shows the pseudo-code used to generate the file that BILIN accepts.
bilin file previously described.
You can download the bilin file for this HDF-EOS2 file
here.
With the generated file, the description file can correctly associate
scientific data with coordinate variables. Type the following commands
at the GrADS prompt in order to draw a shaded plot with the description
file for a polar stereographic projection.
set mproj nps indicates that the plot is drawn over a
north polar stereographic map. GrADS 2.0a2 gave an error saying
that HDF SDS was not fully implemented. This problem disappeared in
GrADS 2.0a3, and we could draw the Snow Depth image shown in Figure 12.
We believe GrADS can handle all types of projections in HDF-EOS2 grid,
as long as the user provides the file that BILIN needs.
For swath, this can be more difficult because HDF-EOS2 does not have
a function similar to GDll2ij to calculate the index for
swath data. Users need to interpolate EOS Swath to regular
gridded data to obtain the array index for each grid point.
The discussion of this topic is beyond the scope of this document.
For more information about using the BILIN option to draw
a plot with GrADS, please read Appendix 5.3.
Besides BILIN, GrADS provides a convenient
method to generate plots for files having the north polar stereographic projection
or the south polar stereographic projection. Several parameters are
required by GrADS to generate plots for files having these projections.
However, this method may not be straightforward for handling HDF-EOS2 files because HDF-EOS2 and GrADS use polar stereographic projection differently. As of this writing, the authors still cannot draw the correct plot with this method.
This case is very similar to the case described in
Section 4.3.2.4.1
because PDEF BILIN is not format-dependent.
The file that the BILIN option requires is exactly the same
as the one used in Section 4.3.2.4.1.
However, the description file needs to be slightly changed as shown in
Figure 13.
DTYPE is different. Opening this description file with the
open command produces the same plot as shown in
Figure 12.
Although GrADS does not support HDF5, it does support the netCDF-4 classic model. This subsection explains what the netCDF-4 classic model-compliant HDF5 file is and how to get it from an HDF5 file.
A netCDF-4 classic model-compliant HDF5 file strictly follows the netCDF-3 data model. It has three requirements. The first requirement is that each dimension of an HDF5 dataset associated with a dimension dataset and the dimension should be located under the same or ancestor groups of the HDF5 dataset it is associated with. For this reason, a netCDF-4 classic model-compliant HDF5 file is a netCDF-4-compliant HDF5 file. The second requirement is that all HDF5 datasets should be defined only under the HDF5 root group. The third one is that all HDF5 datatypes should be HDF5 atomic datatypes.
Most netCDF-4-compliant HDF5 files contain several groups, which causes them not to conform to the netCDF-4 classic model. Although it is tedious, it is not impossible to manually convert a netCDF-4-compliant HDF5 file with a non-classic model into one with a classic model.
One way to accomplish this is to use ncdump and ncgen, which are part of the netCDF-4 package. One can use ncdump to generate a text file from a netCDF-4 file, which is shown in Figure 14. With this text file, one can use any text editor to remove groups, as shown in Figure 15. One may also need to rename some variables if their names are used in multiple groups. Then, the modified text file can be used as input to ncgen, which generates a netCDF-4 file. The output of ncgen can be read by GrADS.
This section explains how users can retrieve dimensions associated with the given
variable using
HDF View . Both dimension names and dimension sizes are
necessary to write a description file used by
. Both dimension names and dimension sizes are
necessary to write a description file used by
xdfopen command.
Opening an HDF-EOS2 file shows the hierarchy of the opened file. For example, users will see Figure 16 when the AMSR-E file that contains the rainfall accumulation data is opened.
To retrieve dimensions associated with RrLandRain variable, the user can click the right button on RrLandRain variable in the tree and choose "Show Properties". That command shows detailed properties of RrLandRain variable as shown in Figure 17.
The user can find dimension information from the row marked as "Dimension Size(s)" located in the middle of the window. The string can be interpreted as follows:
YDim:MonthlyRainTotal_GeoGrid.
XDim:MonthlyRainTotal_GeoGrid.
XDEF and YDEF in the description file.
This section provides more information about the PDEF BILIN
option with a concrete example.
Suppose that a data field, Field1, has 3×3 values,
and they are measured at
(20˚S, 30˚W), (20˚S, 0˚), (20˚S, 30˚E);
(0˚, 30˚W), (0˚, 0˚), (0˚, 30˚E);
(20˚N, 30˚W), (20˚N, 0˚), (20˚N, 30˚E).
For this data field, one can write a description file containing the following statements:
XDEF and YDEF specify that the rectilinear
latitude/longitude grid has 3×2 locations:
(20˚S, 30˚W), (20˚S, 0˚), (20˚S, 30˚E);
(20˚N, 30˚W), (20˚N, 0˚), (20˚N, 30˚E).
Figure 20
shows this configuration; nine circles describe where nine elements of
Field1 were measured, and six rectangles describe a rectilinear
latitude/longitude grid defined by XDEF and YDEF.
Note that the setting of a rectilinear grid is up to the users,
and it may have nothing to do with the data field.
For each location of the rectilinear latitude/longitude grid, GrADS must know which element of the data field is related to this location. The related element is specified by its indices for each dimension: i and j. For this example, the following table shows the values of i and j.
| ||||||||||||||||||||||||||||
| Table 2 Locations and indices |
Then, the file that BILIN needs will contain both six values of
i and six values of j, sequentially;
that is, the file will start with 0, 0, 0, 2, 2, 2, 0, 1, 2, 0, 1, 2.
After these values, this file should have additional data for wind rotation values.
For more information, refer to
http://www.iges.org/grads/gadoc/pdef.html .
.
If the rectilinear latitude/longitude grid has a location such as
(10˚S, 15˚E), the values of i and j will be 0.5 and 1.5, respectively.
For this i and j pair, GrADS does the interpolation using data
values from Field1[0][1], Field1[0][2],
Field1[1][1] and Field1[1][2].
Since calculating the value of i and j from a location
is difficult in most cases, the user needs to consider using an HDF-EOS2 API,
GDll2ij, if the file is an HDF-EOS2 grid file. This function returns
i and j based on a location, a projection code and
related attributes. One can easily read the projection code and related
attributes using HDF-EOS2 API
[13][14].